Intrinsically disordered proteins
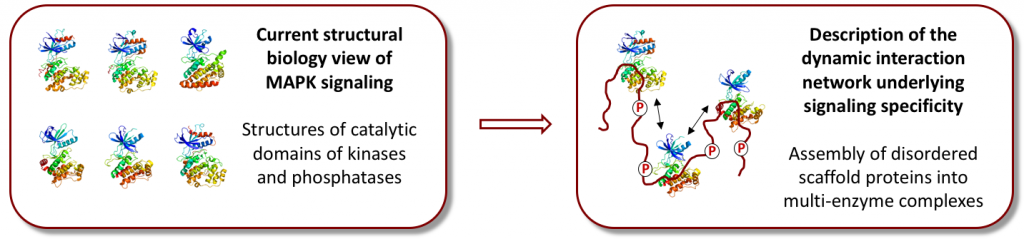
Over the last two decades, it has become increasingly clear that a large fraction (up to 40%) of the proteins encoded by the human genome are intrinsically disordered or contain disordered regions of significant length. These intrinsically disordered proteins (IDPs) play important regulatory roles throughout biology, and they have been intimately linked to a number of human diseases underlining the importance of understanding their interactions and conformational properties at the molecular level. Bioinformatic studies have placed proteins implicated in cell signaling among those with the highest levels of intrinsic disorder in the human genome. Yet, from a structural biology perspective, our current knowledge about signal transmission is essentially limited to crystal structures of folded, catalytic domains of kinases and phosphatases. In our research team we are interested in elucidating the role of intrinsic disorder in the mitogen-activated protein kinase (MAPKs) cell signaling pathways with a particular focus on assembly of scaffolding complexes.
Mitogen-activated protein kinases and the role of scaffold proteins
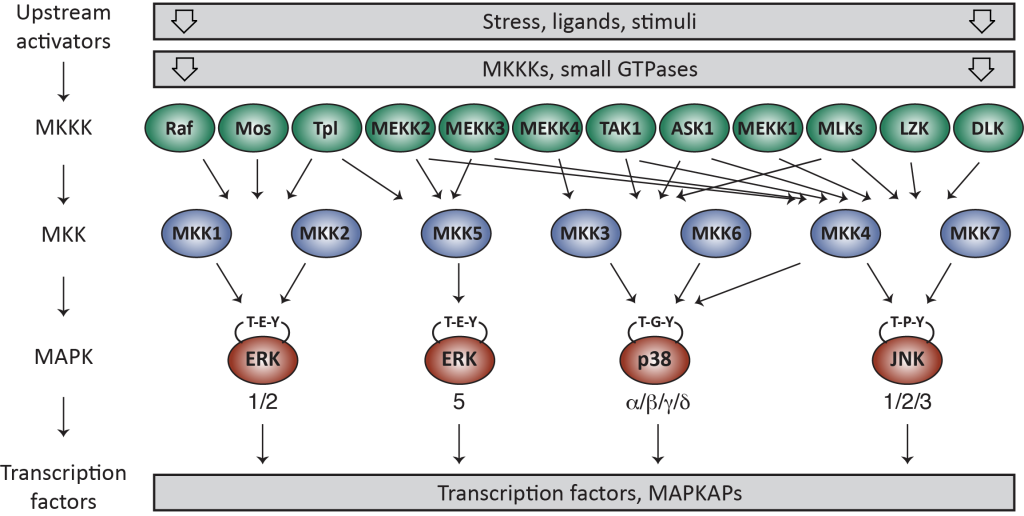
Mitogen-activated protein kinases (MAPKs) are essential components of eukaryotic signal transduction networks that transfer signals from activated receptors on the cell surface to various cellular compartments, notably in the nucleus, where they control cellular processes such as activation of gene transcription, protein synthesis, cell cycle, cell death and differentiation. The MAPK pathways feature three sequentially acting protein kinases making up a signaling module: an MKKK that phosphorylates and thereby activates an MKK, which then activates the MAPK by phosphorylation. The activated MAPK can then phosphorylate downstream substrates such as transcriptional regulators. Four major signaling pathways have been identified: the two ERK (extracellular-signal-regulated kinase) pathways, p38 and JNK.
One of the outstanding questions concerning MAPK signaling is how specificity is regulated at the molecular level. Intrinsically disordered scaffold proteins play crucial roles in ensuring signaling specificity by assembling multiple kinases into dynamic signaling complexes. This assembly leads to sequential activation of the bound kinases, whilst avoiding cross-reactivity with neighboring pathways. Despite the discovery of their role more than 20 years ago, we still have little idea about the molecular mechanisms employed by scaffold proteins. The main reason for this is the dynamic nature of many scaffolds with intrinsically disordered domains covering regions of up to 600 consecutive amino acids. This poses a number of challenges for contemporary structural biology as atomic resolution information is needed of these highly dynamic regions to unravel the molecular and regulatory details that underlie scaffold protein function.
The goal of our research is to obtain a complete mechanistic description of scaffold protein function within the c-Jun N-terminal kinase (JNK) cell signaling pathway. We will reveal how JNK scaffold proteins recruit and discriminate between kinases, how signals are transmitted across the scaffolding complexes and how they are regulated by phosphorylation. This includes mapping of precise binding sites and residence times for kinases and phosphatases on the scaffolds and determination of the affinity, structure and dynamics of individual sub-complexes. In addition, we assemble entire scaffolding and super-scaffolding complexes to reveal the hierarchy of assembly and interplay between kinases and phosphatases and to probe structural, dynamic and allosteric rearrangements of the scaffold proteins during assembly. Finally, we are interested in the role of post-translational modifications where we map phosphorylation sites in the scaffold proteins and study how these modifications regulate interactions with kinases and phosphatases and complex assembly in general.
Methodology and experimental techniques
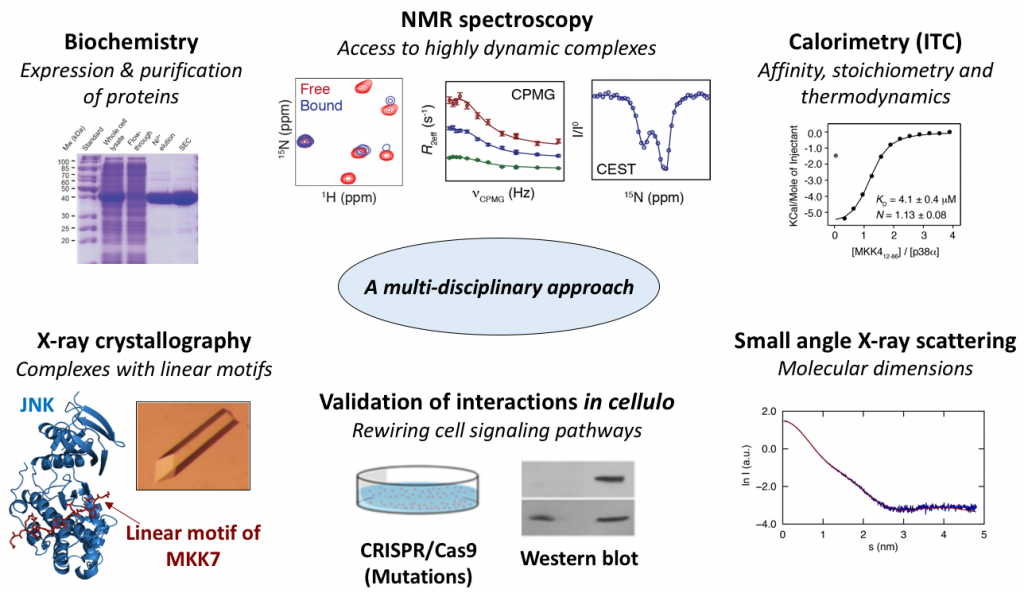
We use a multi-disciplinary approach where the main experimental technique is nuclear magnetic resonance (NMR) spectroscopy that provides atomic resolution information about IDPs. In particular, NMR allows characterization of the structure and dynamics of IDPs and mapping of their interaction mechanisms. We employ a range of different NMR techniques to study the assembly of dynamic complexes including NMR exchange techniques such as chemical exchange saturation transfer (CEST), CPMG relaxation dispersion and off-resonance R1rho experiments that provide insight into conformational exchange processes occuring on time scales ranging from a few milliseconds to tens of microseconds.
We combine our NMR studies with X-ray crystallography in collaboration with Andrés Palencia’s research group (IAB, Grenoble) to obtain structures of kinases and phosphatases in complex with linear motifs of IDPs. The high-resolution structural information allows us to identify stabilizing interactions necessary for complex formation and to design appropriate mutations for testing in cellulo and in vivo.
The structural biology data are combined with biophysical measurements mainly using isothermal titration calorimetry (ITC) that provides information about the affinity, stoichiometry and thermodynamic profile of the IDP complexes.
Last but not least, we validate our interactions in cellulo and in vivo in collaboration with Roger Davis’ research group (University of Massachusetts Medical School). This includes quantification of signaling through scaffolding complexes by mutational analysis of specific residues or entire regions observed to be crucial for complex assembly by using the gene editing tool CRISPR/Cas9. Based on the in cellulo results, we select mutations for further study using physiological testing in mice to probe scaffold protein function in vivo.
Applications and translational aspects of our research
Deregulation of the MAPK pathways is implicated in the development and progression of human cancers, and the MAPKs, therefore, represent promising drug targets. Both the academic community and the pharmaceutical industry have invested in the development of MAPK inhibitors by targeting the ATP-binding pocket. These investments have led to only sub-optimal inhibitors as many of the molecules suffer from a lack of specificity due to the high conservation of the ATP binding pocket across kinases. As a long-term objective of our research, we therefore propose a novel strategy to control MAPK activity i.e. to interfere with the protein-protein interactions mediated by scaffold proteins. Our atomic resolution studies of the mechanism of action of this family of proteins, combined with testing in cellulo and in vivo, will be invaluable for guiding medicinal chemistry in the development of next-generation anti-cancer drugs.
Facilities
We are located at the Institut de Biologie Structurale in Grenoble where we have access to a number of state-of-the-art structural biology facilities. In particular, we benefit from the NMR platform with six high-field spectrometers (3 x 600, 700, 850 and 950 MHz) equipped with cryoprobes for high-sensitivity solution measurements. In addition, we have dedicated wetlab facilities for cloning, expression and purification of proteins as well as access to a number of research platforms through the ISBG (Integrated Structural Biology Grenoble). Importantly, this includes biophysical techniques for quantitative measurements of protein-protein interactions (isothermal titration calorimetry, thermophoresis, surface plasmon resonance). For X-ray crystallography, we make use of the crystallization platform located at the EMBL, Grenoble that offers completely automated crystallization screening of proteins under various conditions and for diffraction of crystals we have priviliged access to the beam lines at the ESRF.

Key publications
(1) L. Marino Perez, F. S. Ielasi, L.M. Bessa, D. Maurin, J. Kragelj, M. Blackledge, N. Salvi, G. Bouvignies, A. Palencia*, M.R. Jensen*. Nature (2022) 602, 695-700. “Visualizing protein breathing motions associated with aromatic ring flipping”
(2) Jaka Kragelj‡, T. Orand‡, E. Delaforge, L. Tengo, M. Blackledge, A. Palencia, M.R. Jensen*. Biomolecules. (2021) 11, 1204. “Enthalpy–entropy compensation in the promiscuous interaction of an intrinsically disordered protein with homologous protein partners”
(3) R. Schneider, M. Blackledge, M.R. Jensen*. Curr. Opin. Struct. Biol. (2019) 54, 10-18. “Elucidating binding mechanisms and dynamics of intrinsically disordered protein complexes using NMR spectroscopy”
(4) S. Milles, N. Salvi, M. Blackledge, M.R. Jensen*. Prog. Nucl. Magn. Reson. Spectrosc. (2018) 109, 79-100. “Characterization of intrinsically disordered proteins and their dynamic complexes: From in vitro to cell-like environments”
(5) E. Delaforge, J. Kragelj, L. Tengo, A. Palencia, S. Milles, G. Bouvignies, N. Salvi, M. Blackledge, M.R. Jensen*. J. Am. Chem. Soc. (2018) 140, 1148-1158. “Deciphering the dynamic interaction profile of an intrinsically disordered protein by NMR exchange spectroscopy”
(6) J. Kragelj, A. Palencia, M. Nanao, D. Maurin, G. Bouvignies, M. Blackledge*, M.R. Jensen*. Proc. Natl. Acad. Sci. U.S.A. (2015) 112, 3409-3414. “Structure and dynamics of the MKK7-JNK signalling complex”
